Installing Software on MacOS

For screen-shot slides, click here
For the YouTube narration, click here
This is the software installation walkthrough for those whose computers are running MacOS. Based on our pre-course interest form, you make up a solid proportion of the course participants.
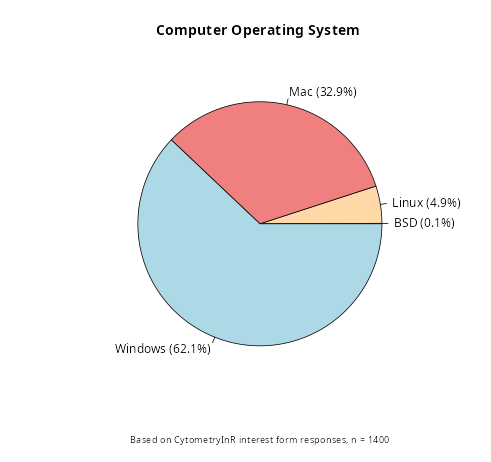
Background
In this walkthrough, we will have you install the following pieces of software:
R website : The programming language we will be using throughout the course.
Positron : The integrated development environment (IDE) in which we will open, modify and run our code.
Git : The version control software that will allow us to track changes to our files. You may or may not already it on your computer depending on whether Xcode Command Line Tools is installed.
Please note, the installation steps detailed below were carried out on a Mac with the older older pre-2020 x86 Intel chips, if you are running a Mac with the newer ARM (M1 or greater) chips, the installation instructions will differ some of the options you selecct during the download. If you encounter major differences in installation, please take screenshots and open an issue so that we can update the documentation for your architecture.
Getting Started
Installing R
To get started, first navigate to the R website. Once there, click on Download R option towards the top of the page.
On the next screen, you will need to select a mirror from which to download the software from. You can either select the closest geographic location (which may be faster) or alternatively just select the Cloud option which should redirect you.

You will then select your Operating System, in this case, macOS

Next, you will need to select the appropiate download based on your computers architecture. On newer Macs (containing M1+ chips) this would the arm64 option on the center left of the screen. For the older Intel (pre-2020) Macs, you would select the x86_64 option. If you are unsure, check your About This Mac tab

After the download has completed, launch the installer
Proceed through the Read Me
You will then be prompted to acccept the software license (which is the free copyleft GPL2 license, which we will learn about later in the course).
Next, you will need to navigate through several pages, keeping the defaults.
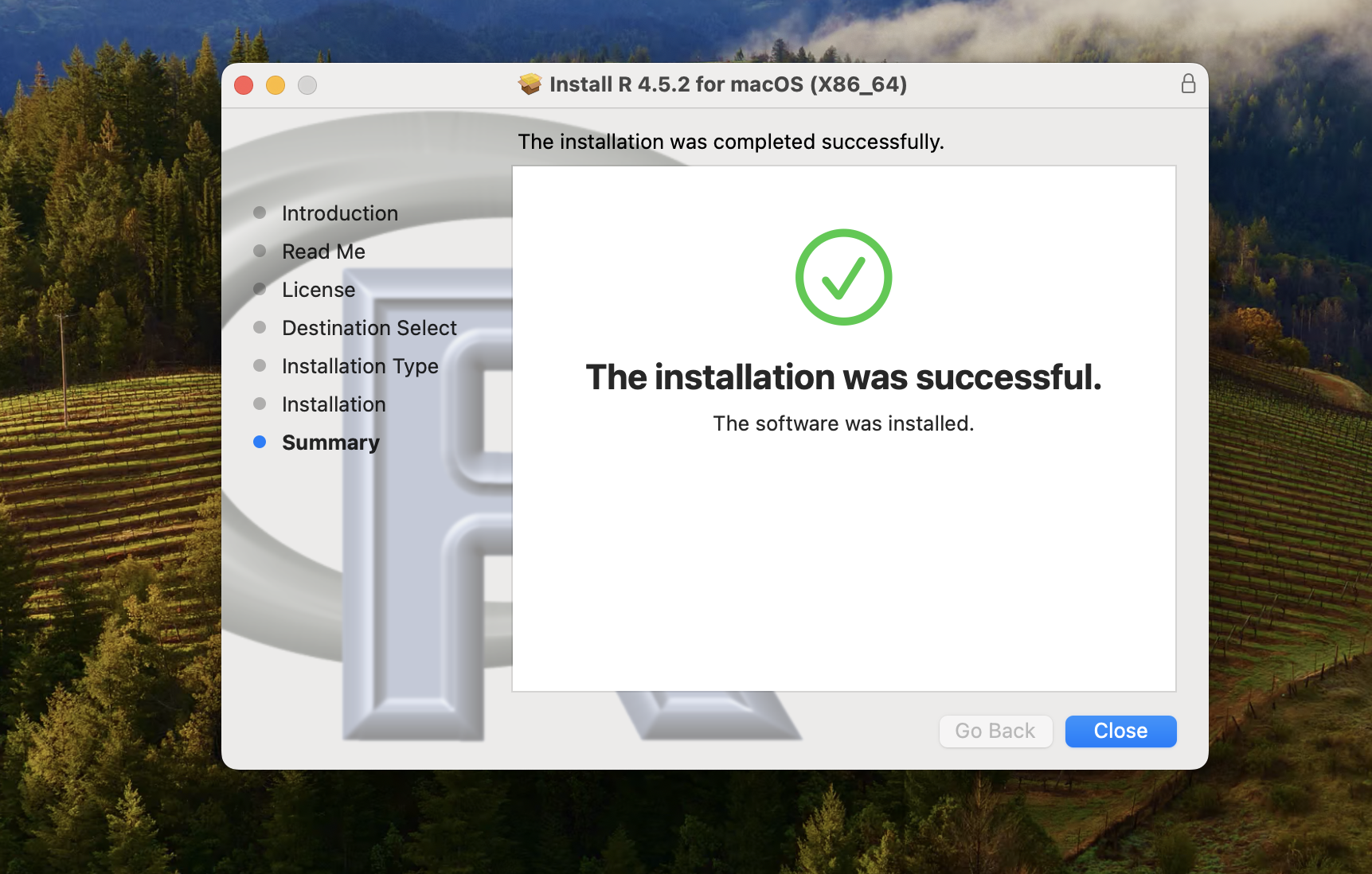
And with any luck, you should see that the installation was successful.
Xcode Command Line Tools
Depending on your version of macOS, you may or may not already have Git installed. The reason for this is it is included within the Xcode Command Line Tools.
Often, when opening Positron for the first time, you will have a popup along the lines of “Git requires command line developer tools. Would you like to install the tools now?”. If you encounter this, select Install. In addition to installing Git, these command line developer tools will also install the the equivalent of the Rtools for Windows, so you will need these tools regardless.
Unfortunately, I had already installed these previously on my Mac, so I was not able to get the necessary screenshots for this documentation. If you encounter this popup on your installation, please take screen shots of each step, and reach out so that I can appropiately update this documentation.
In case the popup doesn’t trigger, and you need to install Xcode Command Line Tools yourself, please see these instructions
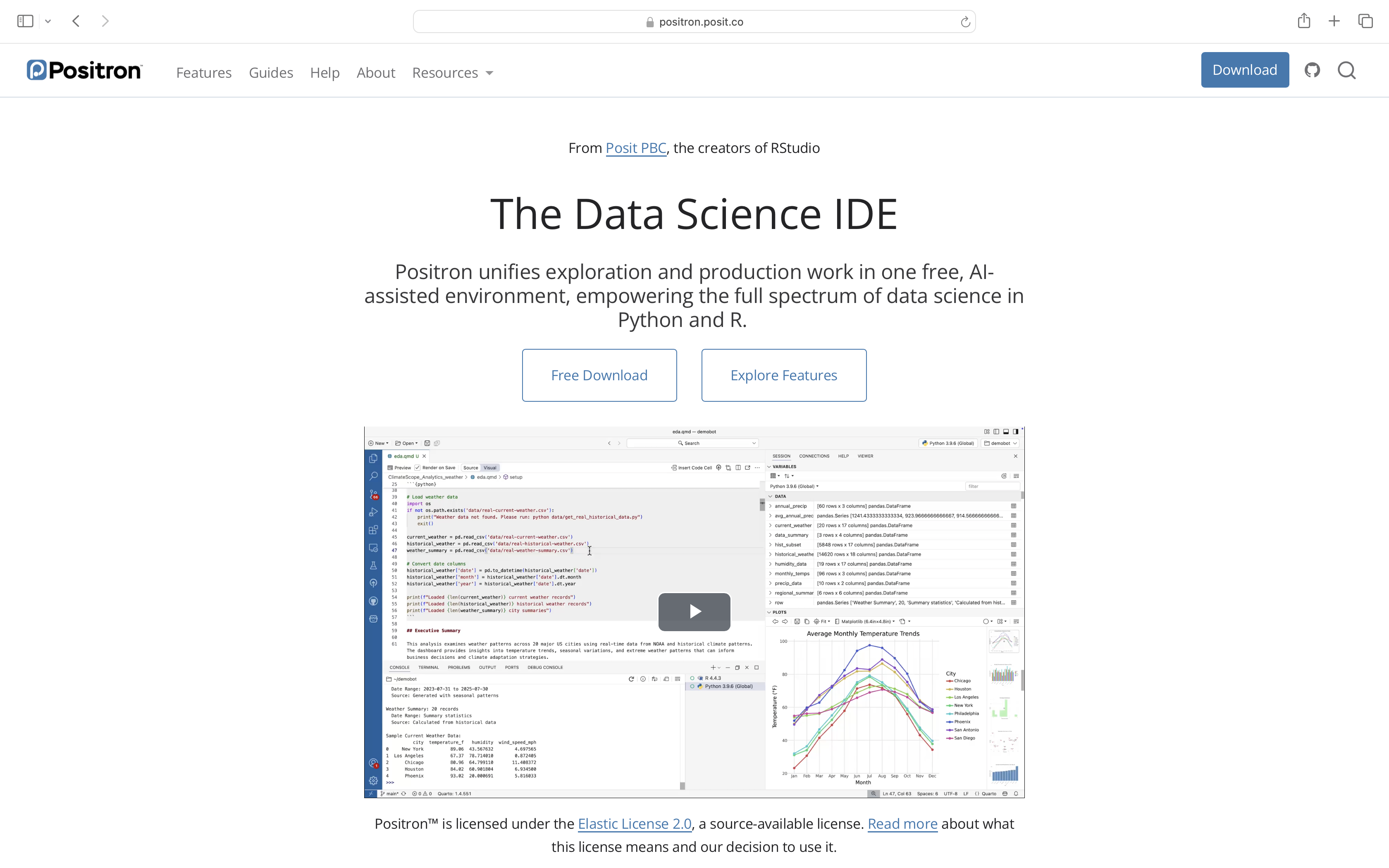
Install Positron
Finally, you will install Positron. It is an integrated development environment (IDE) in which we will open, modify and run our code throughout the course.
First, navigate to their homepage, and select the blue Download option button on the upper-right.
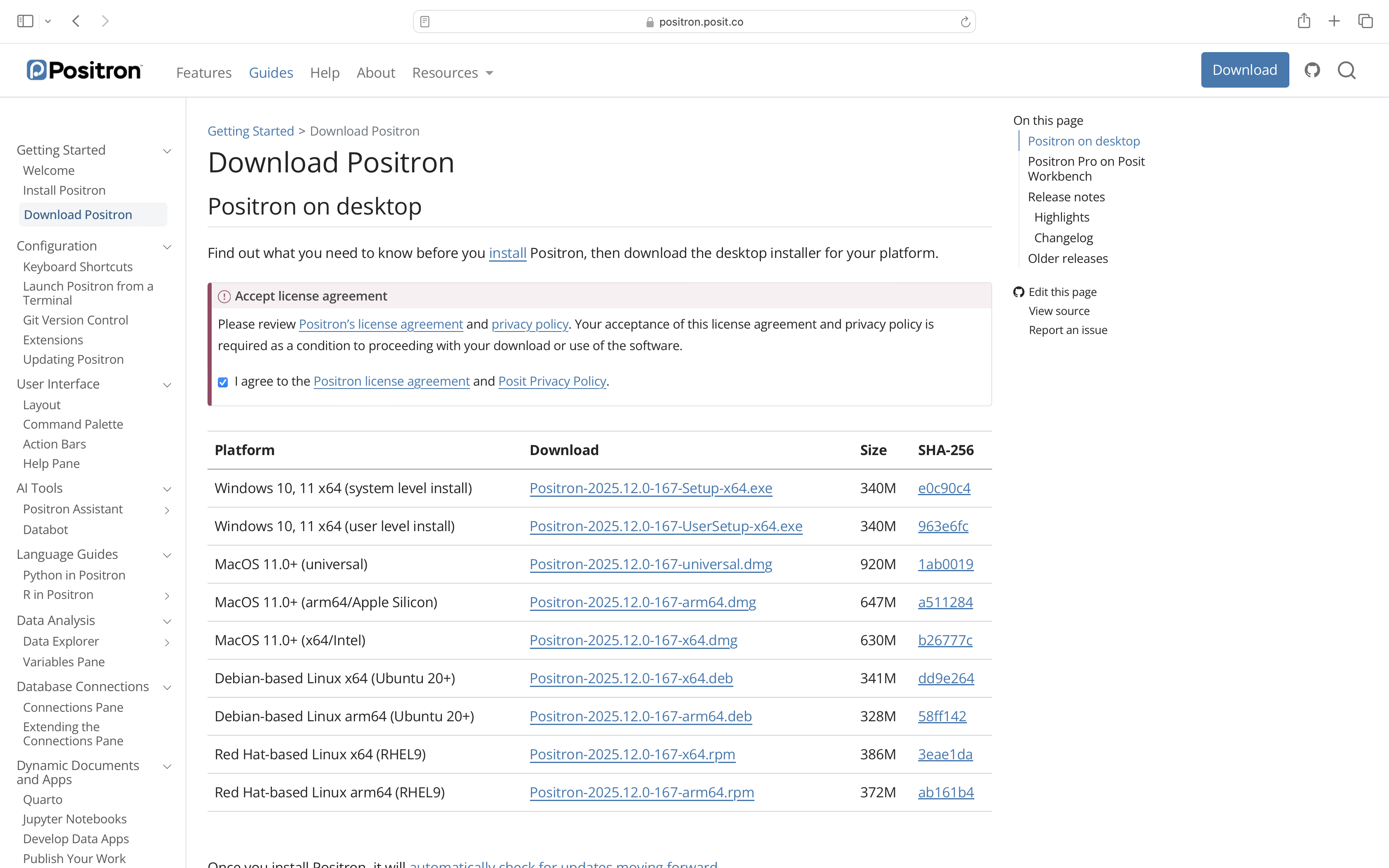
You will then need to accept the Elastic License agreement to use the software (we will cover this source-available license type and what it does later in the course).
With the license accepted, you will be able to select your operating system and the relevant installer depending on whether you are on an M1+ (ARM) or older Intel (x86) Mac.
Once the Download completes, proceed to install the package as you normally would for any other program.
Connecting GitHub and Positron
Now that you have all the software installed, we need to make sure that everything is communicating properly with each other. To do this, we will start by making sure your GitHub account and Positron are connected.
When opening Positron for the first time, you will encounter the following view:
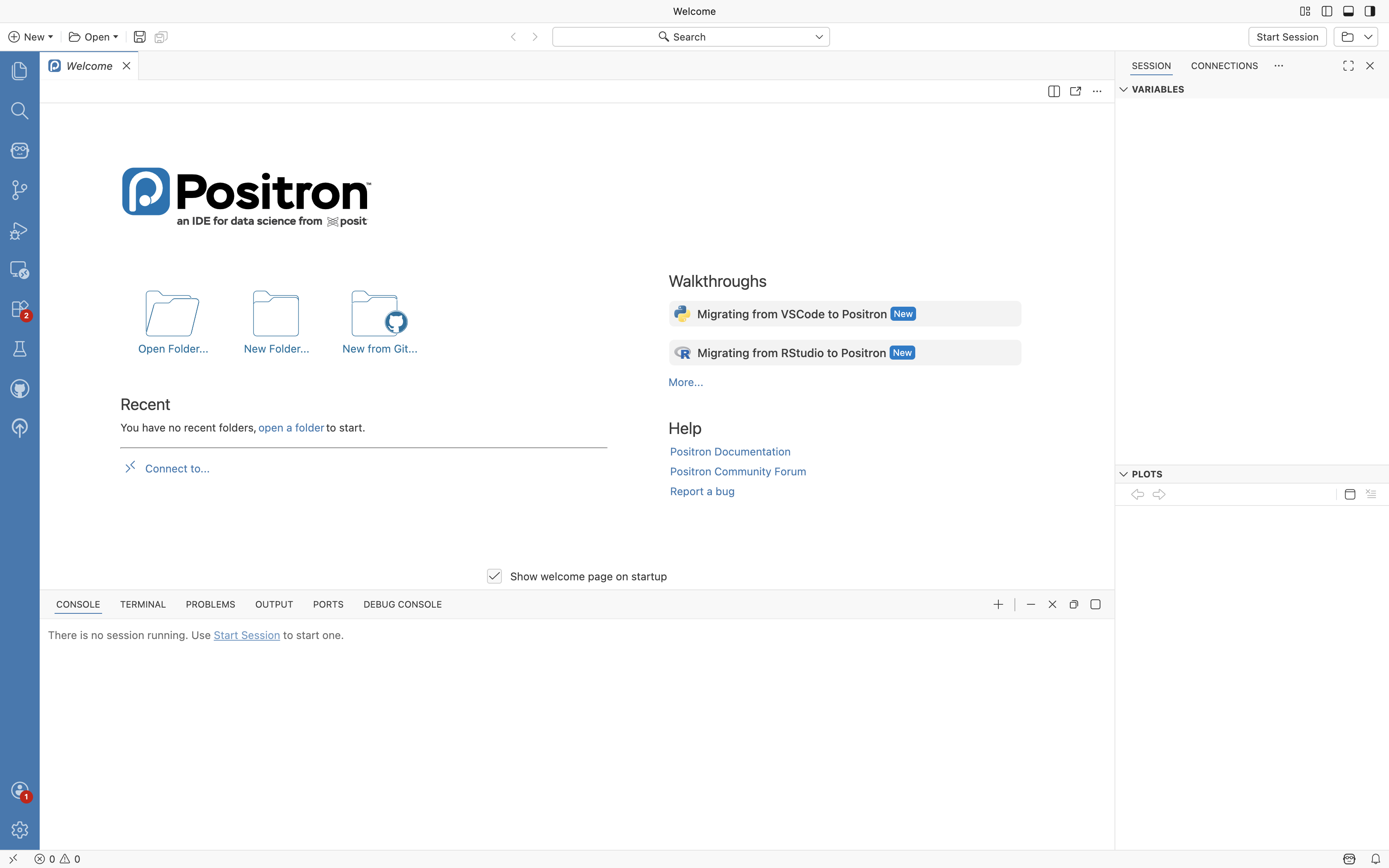
We will go over the various aspects of using Positron in the next section. For now select the “Start Session” button on the upper right, and then select the most recent/up-to-date R version.
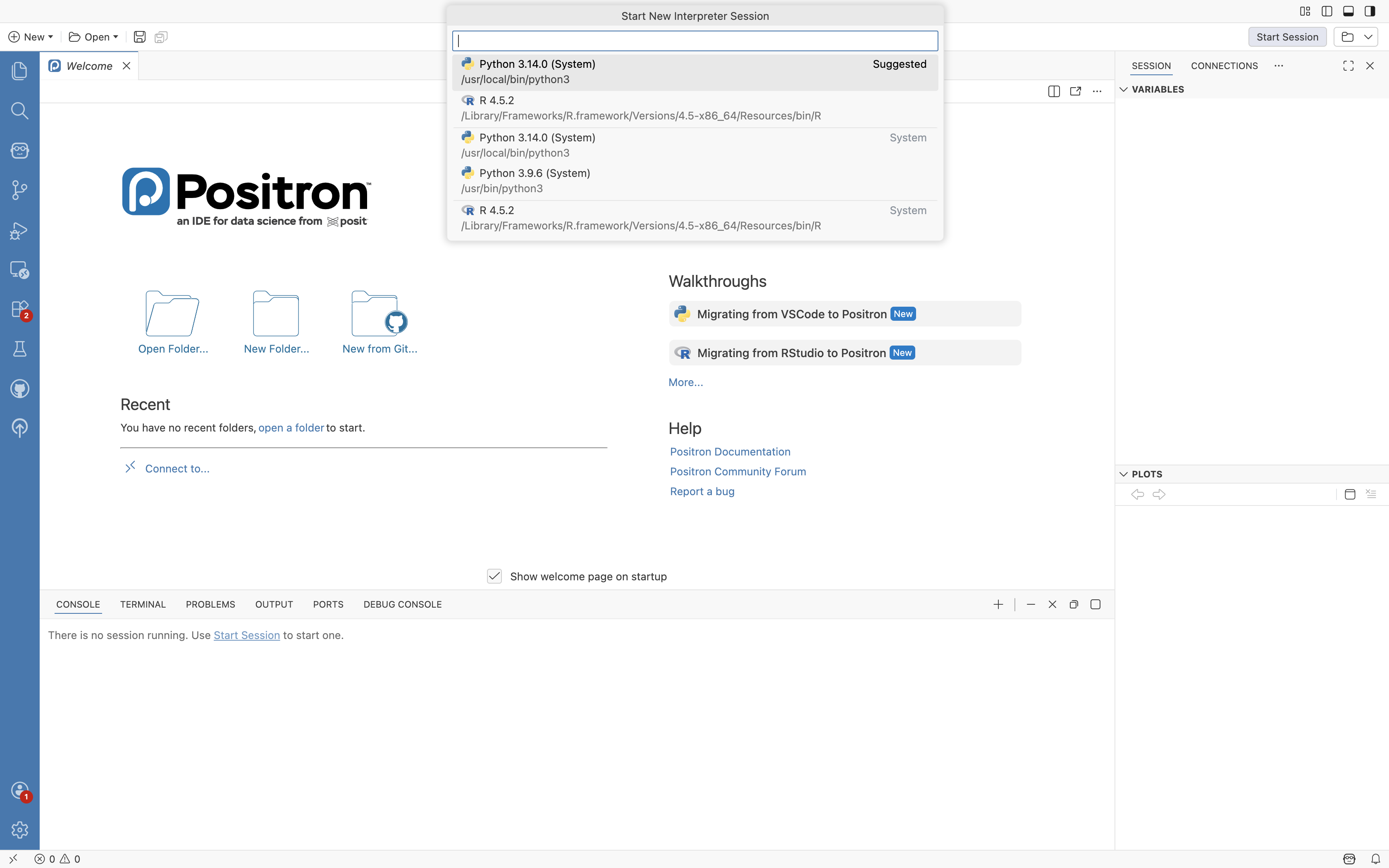
R will proceed to activate, and you will see a message appear in the bottom center window (ie. console)
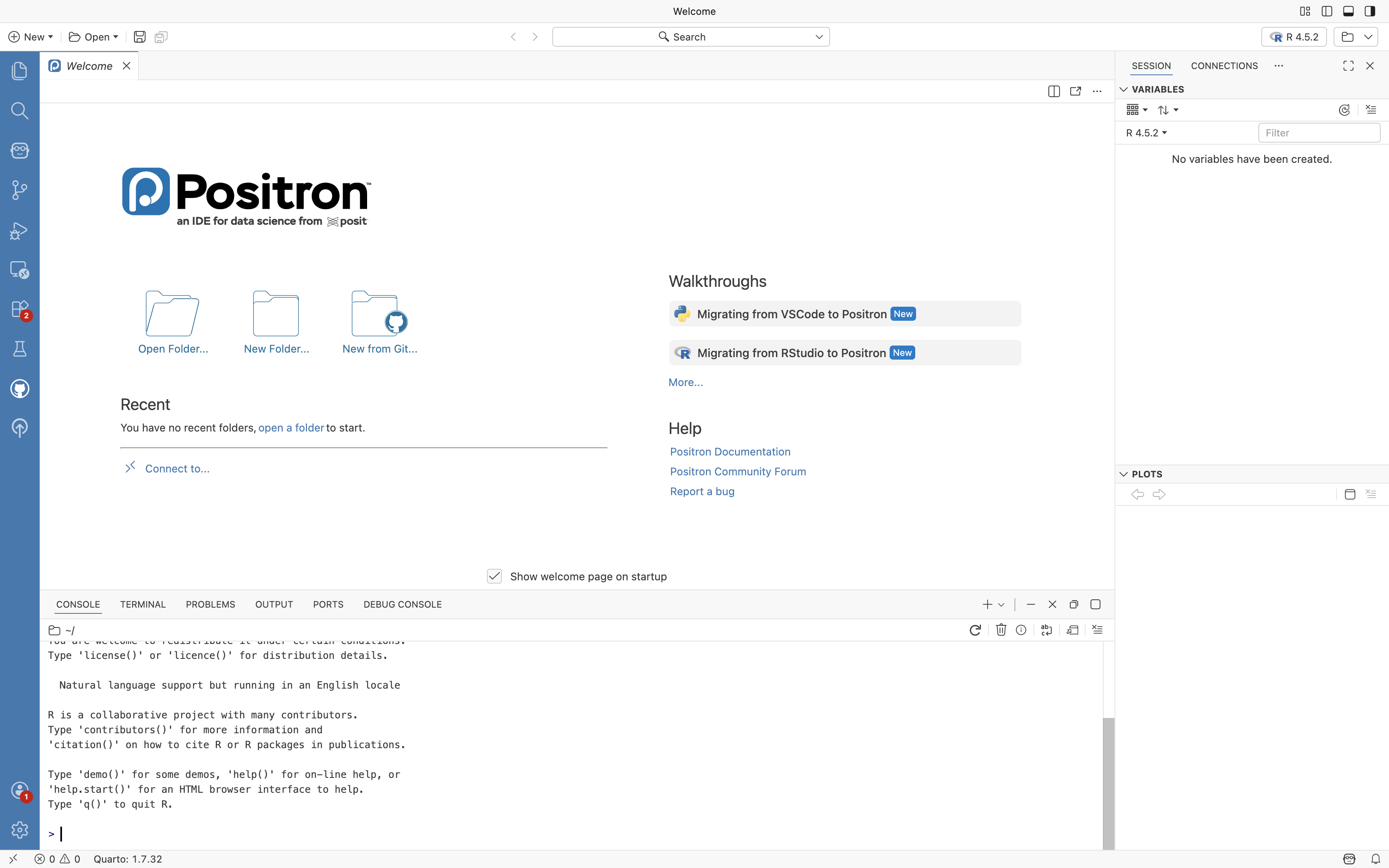
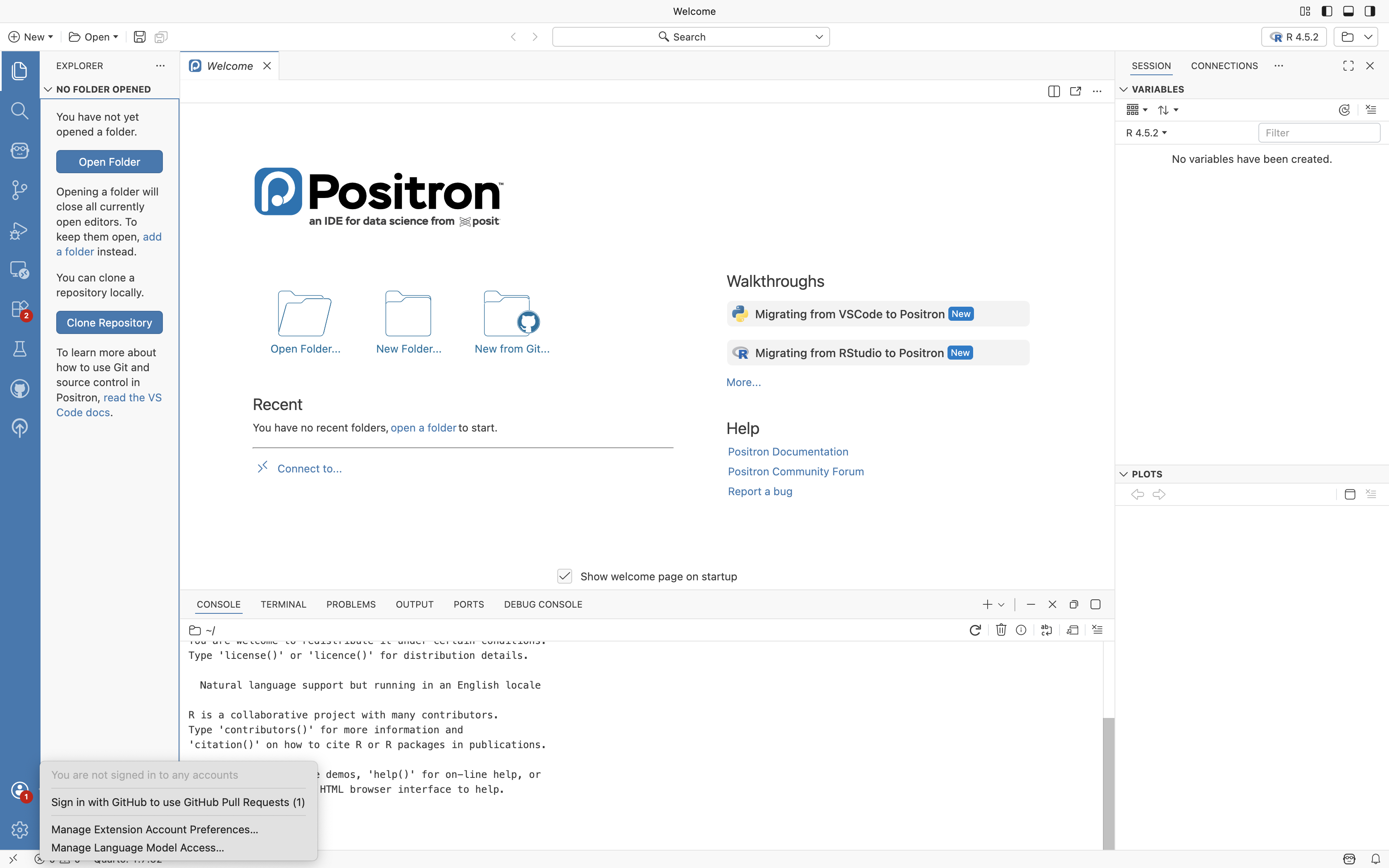
Congratulations, we now know R is working correctly. Next up, connecting your GitHub account to Positron. To make this process easier, first log into your GitHub account from your preferred web browser.
Once this is done, return to Positron, and click on the profile Icon and select “Sign with GitHub” option.
Positron will provide a popup containing a code you will need to give to GitHub to set up authentication. Proceed to click on the “Copy & Continue to GitHub” button.

If you are already signed in, it will ask which account you want to connect Positron with. Select your GitHub account.
You will then be prompted to paste the one-time code provided by Positron. Once pasted, click on Continue.
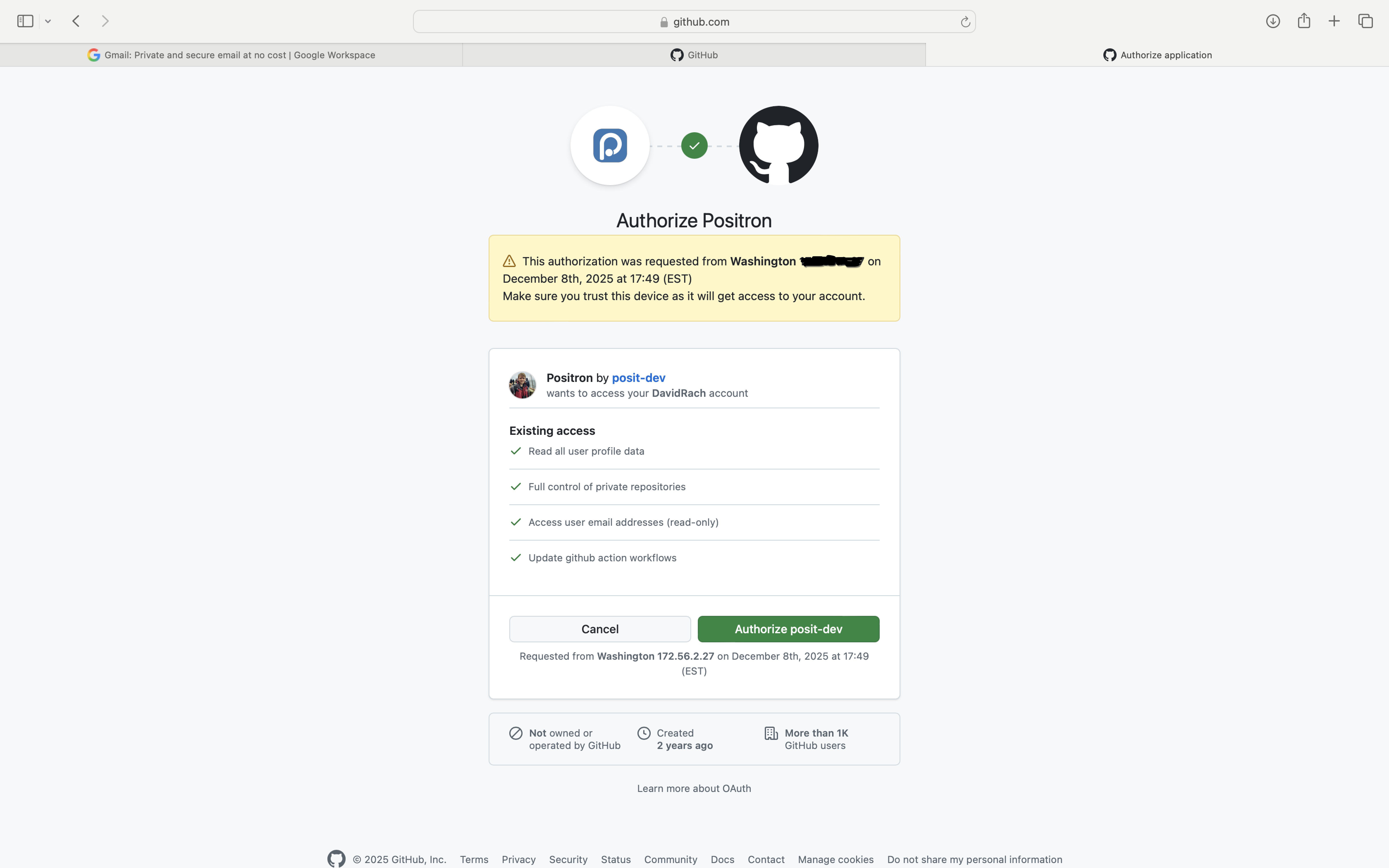
And select “Authorize posit-dev” to authorize Positron to interact with your GitHub Account.
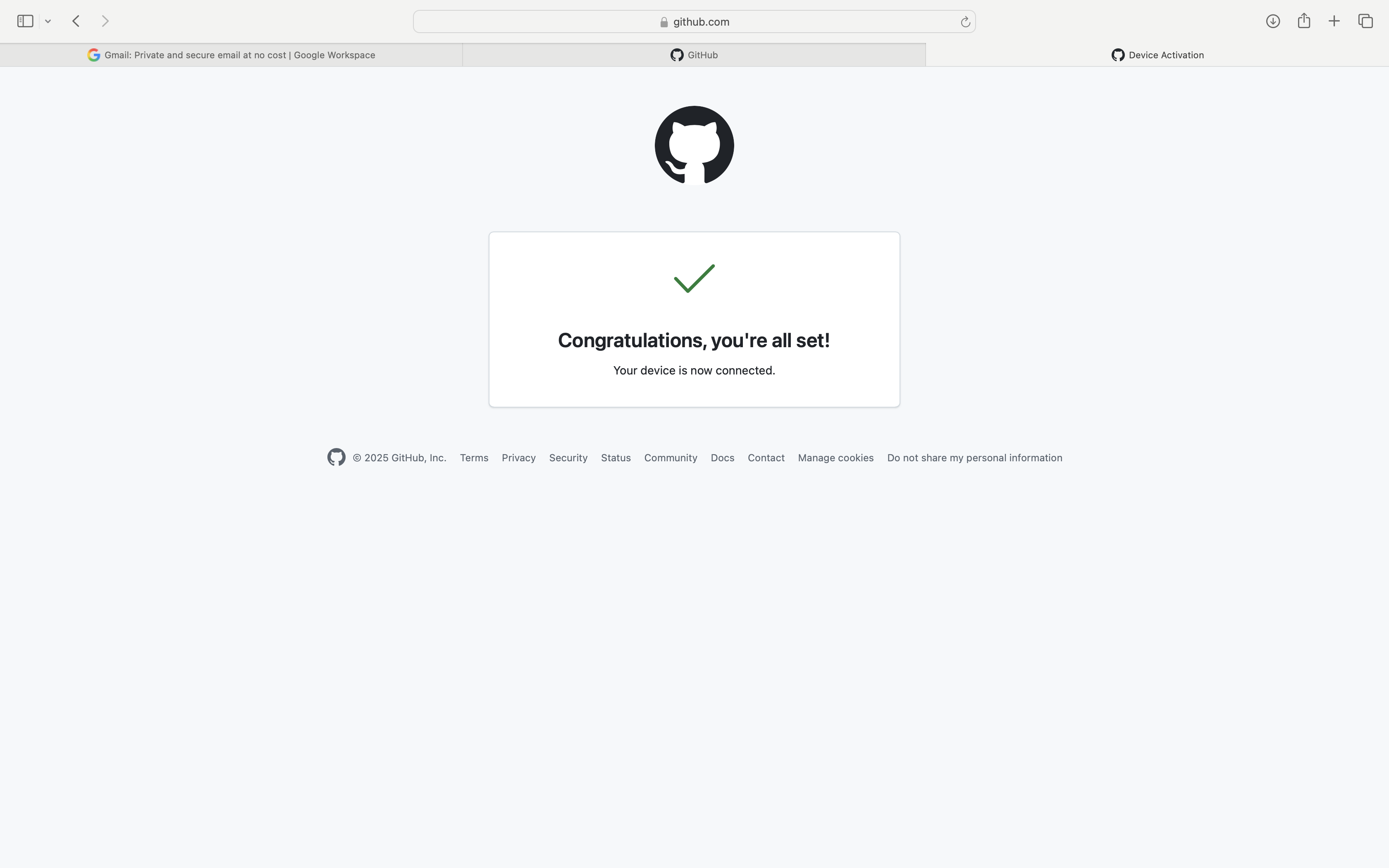
And if all works out, you should see the following succes page
Returning to Positron, check the profile tab on the lower left and verify that it now shows your GitHub account. If it does, congrats! If not, close Positron, check the tab again, and repeat the above one-time code steps if needed.
New Folder from Git
With GitHub and Positron now connected, we will bring your two repositories (the README and the fork of CytometryInR) from GitHub to your local computer.
To this, first navigate to the Folder icon on the upper-right, and select the “New Folder from Git” option
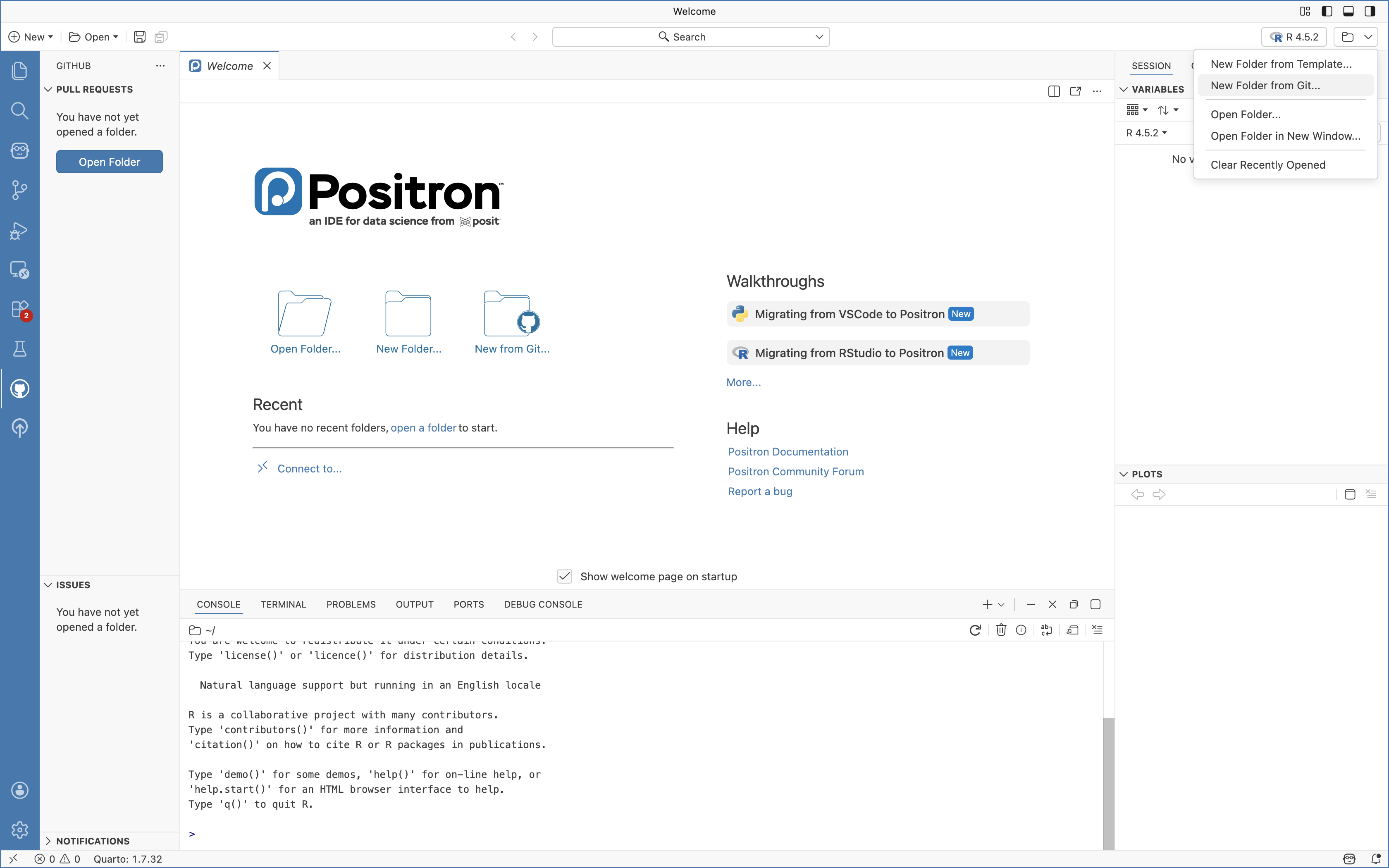
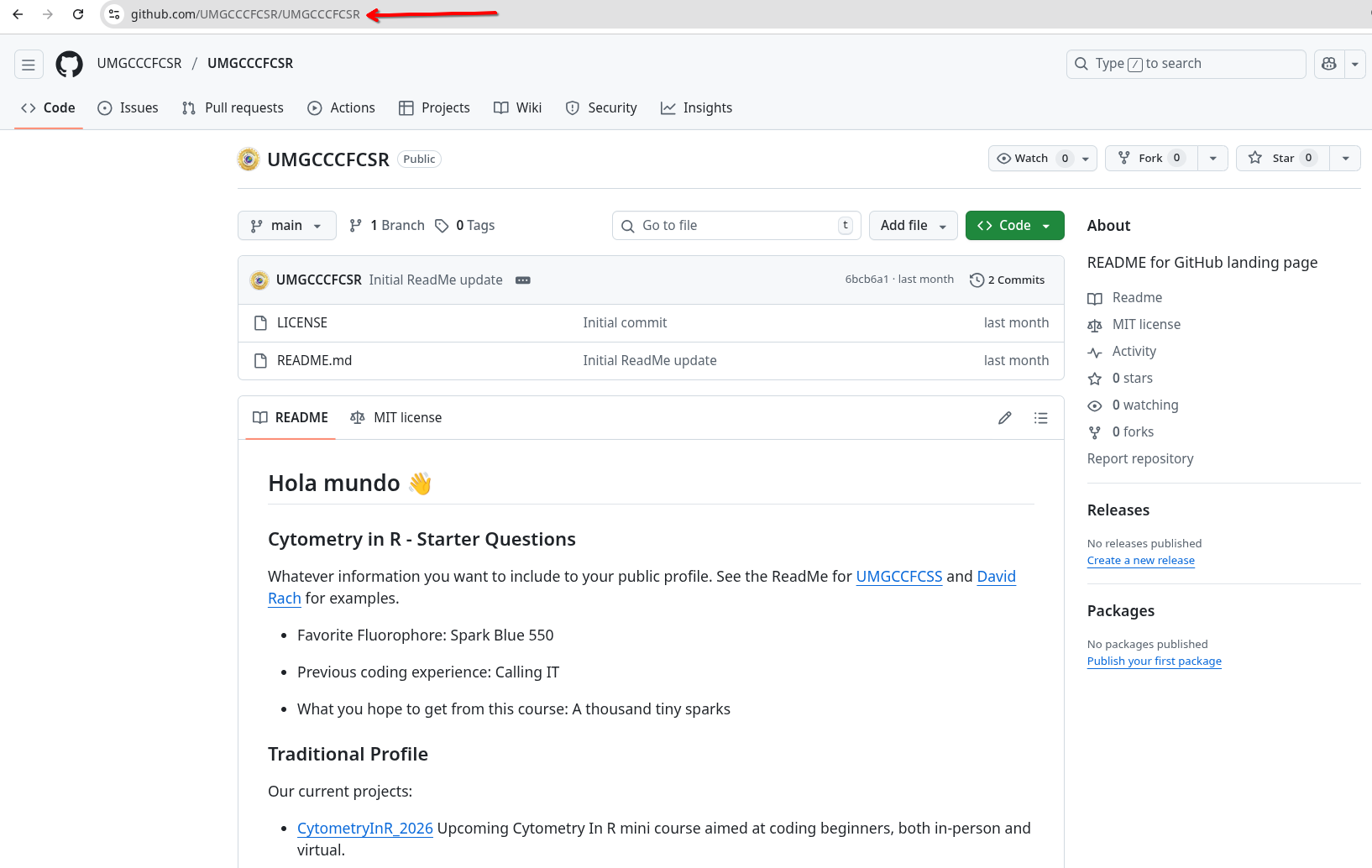
Lets first bring in your forked version of the Cytometry in R repository. Depending on your GitHub username, you would either need to copy the URL from your browser, or swap in your username in place of UMGCCCFCSR in “https://github.com/UMGCCCFCSR/CytometryInR”
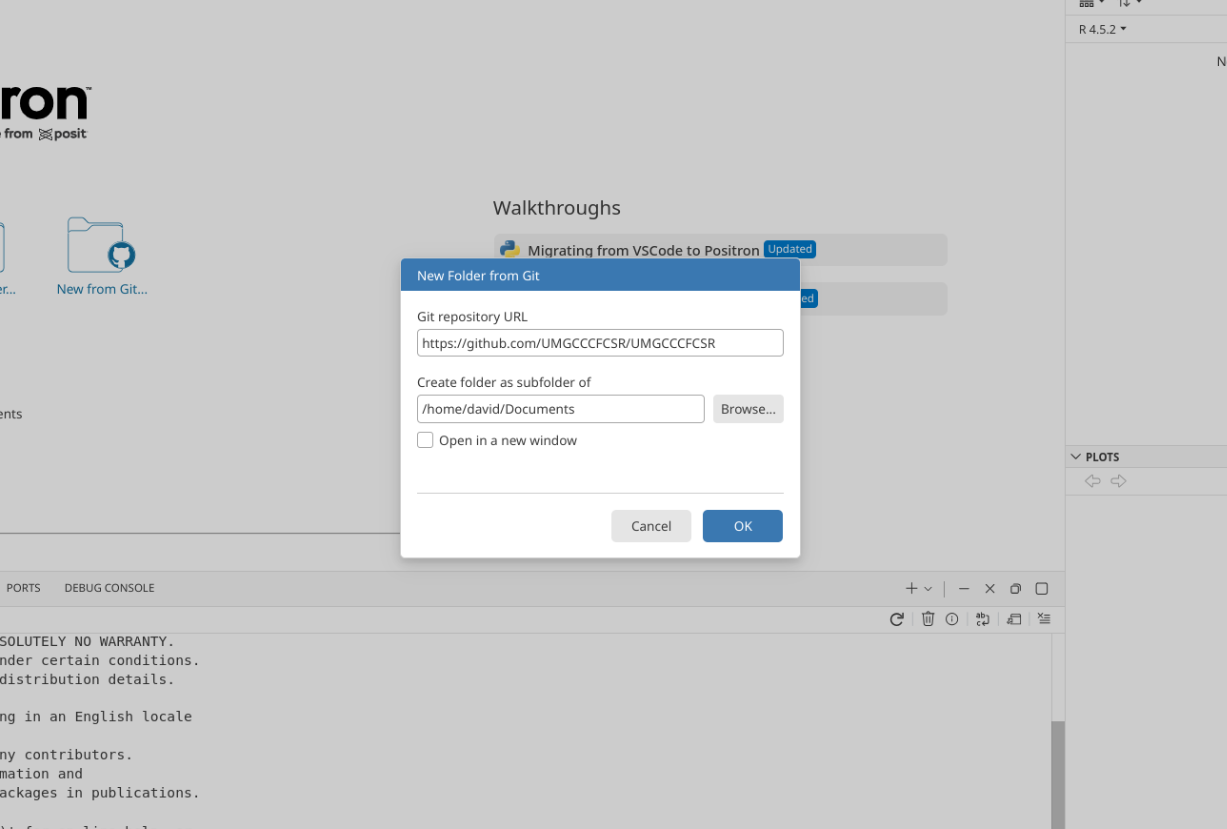
You will also be asked for a location to store your project folder. It is best to save the folder locally (avoid using OneDrive or other cloud locations for now), as it makes things easier to save or modify without running into permission issues. For most of our course examples, we will be saving our Project Folders under the Documents Folder.
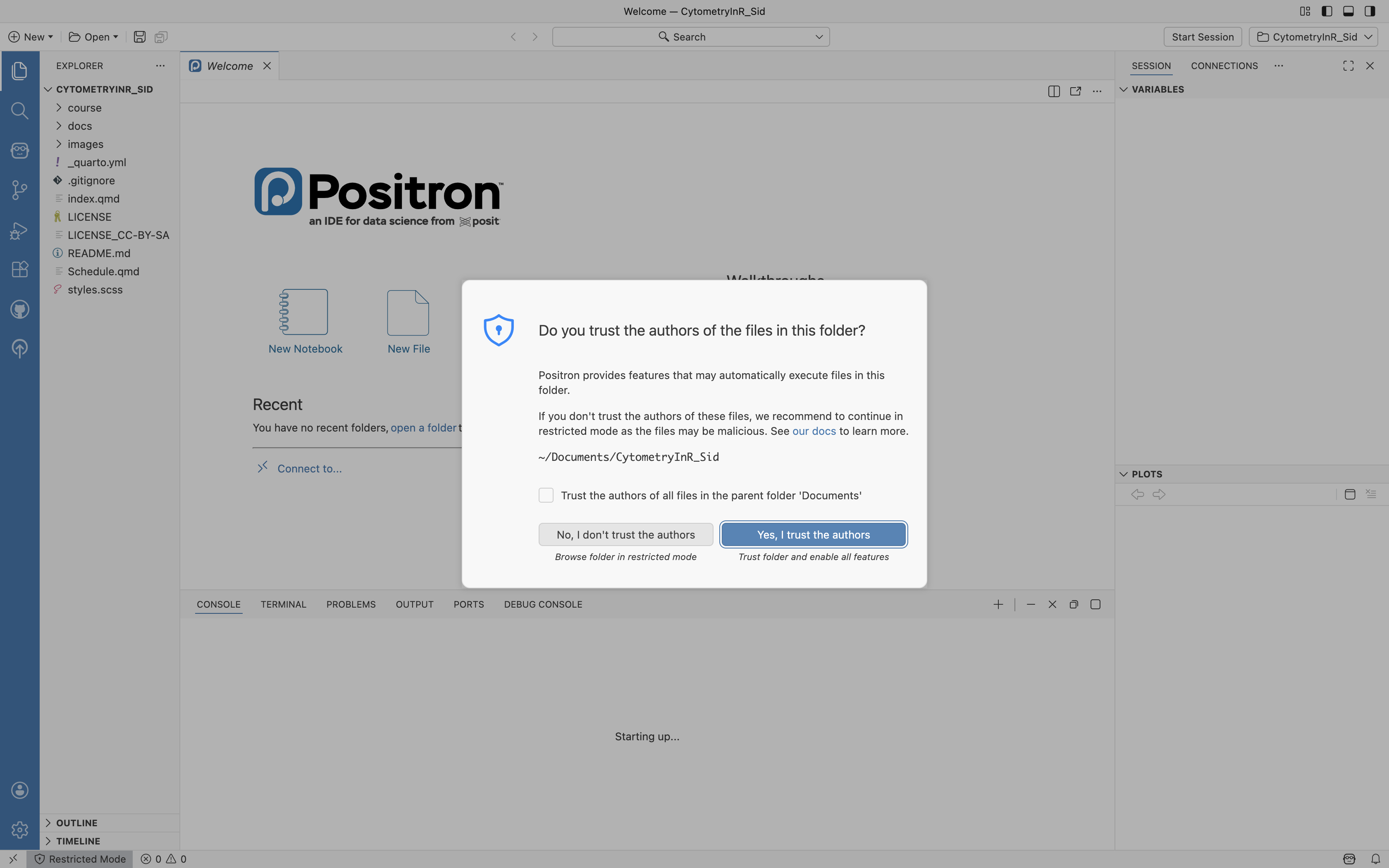
You may encounter a pop-up asking whether you trust the authors for that particular folder. For your own projects, you are the author, so go ahead and select Yes. Selecting No is a good precaution when bringing in a random GitHub project that you haven’t had time to properly vet.
And with that done, you should now see the forked Cytometry in R course contents appear within Positron.
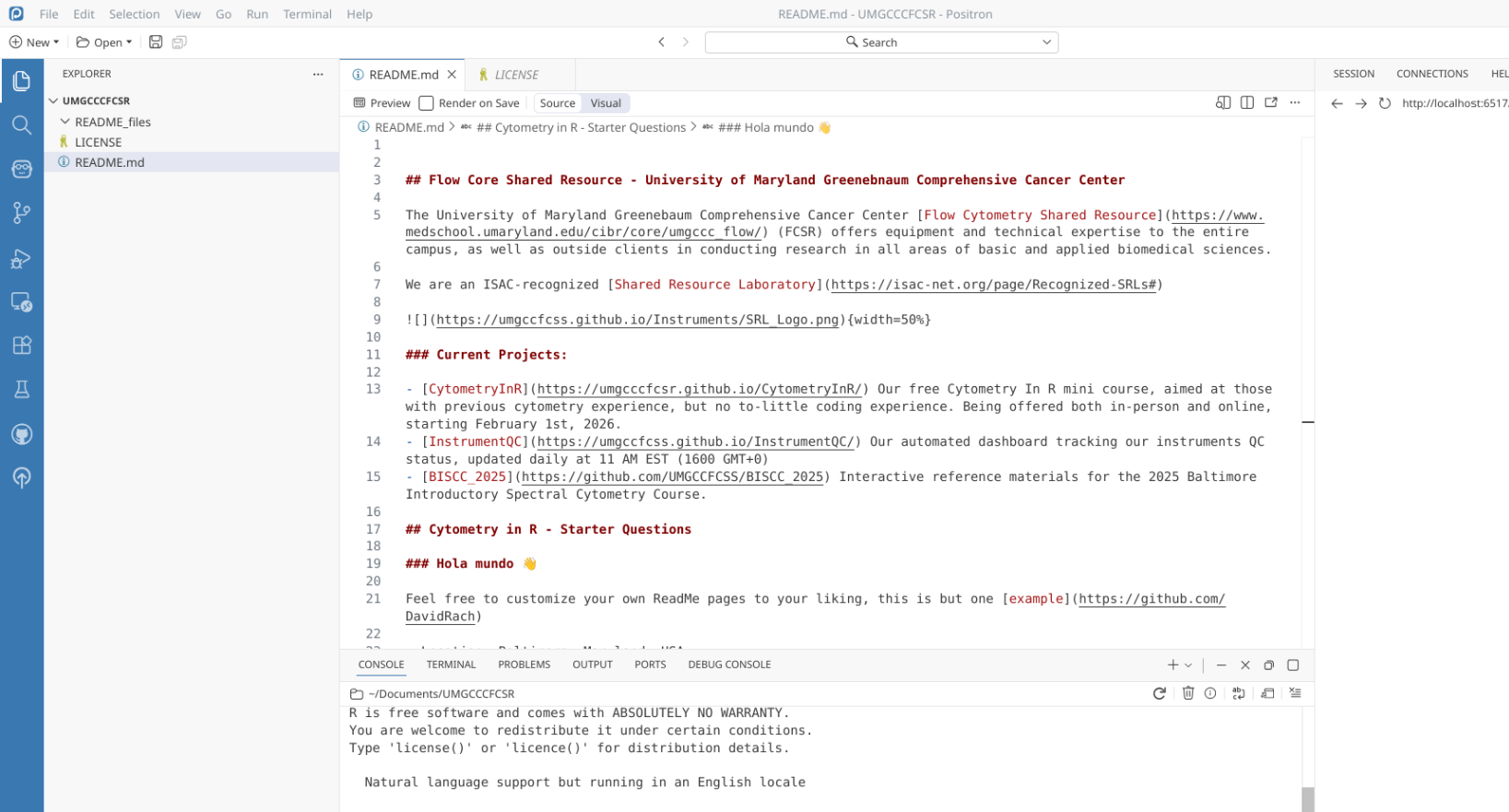
We will now repeat the process to bring in youir GitHub profile’s ReadMe repository. Depending on your GitHub username, you would either need to copy the URL from your browser, or swap in your username twice in place for each UMGCCCFCSR: “https://github.com/UMGCCCFCSR/UMGCCCFCSR”
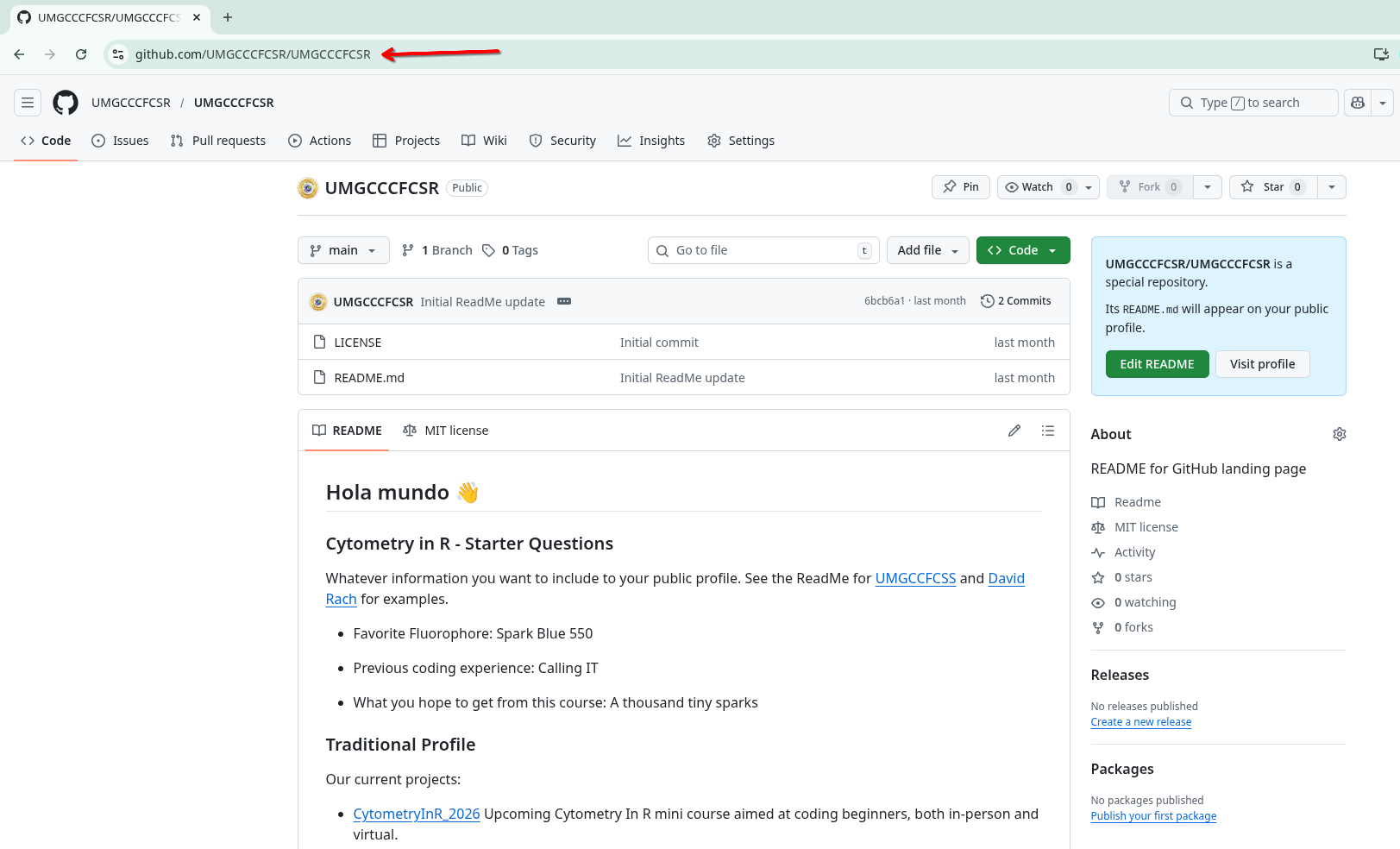
After pasting the URL, and selecting the folder to save your project folder to (Documents in our case), select OK.

And after a quick import you should now see your ReadMe repository files available and ready to be modified on your local computer.

Wrap-up
In this walkthrough, we installed R, Positron, and ensured that Git/Command Line Tools were active. We also made sure to connect Positron and your GitHub account, and brought copies of your repositories into local project folders. With this, you now have on your workstation the major pieces of software needed for the course.
In the next section, we will delve into how to use Positron, and explore more how to use the connection you set up with GitHub to send changes from your local repositories back to GitHub.
Additional Resources
How to Install Software on a Mac
Riffomonas Project: Installing R, RStudio, and packages for Windows and Mac OS X (CC077)



